|
|
Overview
| Common name |
evenia jointvetch |
 |
| Full Name |
Aeschynomene evenia C. Wright |
| Genus |
Aeschynomene |
| Species |
evenia |
| Family |
Fabaceae |
| Subfamily |
Papilionaceae |
| Clade |
Dalbergioids |
The legume genus Aeschynomene L. includes approximately 150 tropical and subtropical species, part of them having a semi-aquatic lifestyle. Some hydrophytic Aeschynomene species display unusual symbiotic features such as stem nodulation and the presence of a Nod factor-independent infection process with some Bradyrhizobium strains.
To decipher the mechanisms of this original symbiotic process, Aeschynomene evenia has emerged as a new model legume because of its advantageous genetic and developmental characteristics for molecular genetics. A. evenia (2n=20, 400 Mb/1C) is an autogamous diploid species that is annual or short-lived perennial, consisting of various genotypes.
The sequenced A. evenia genotype is an inbred line produced by IRD from the accession CIAT22838 originating from Malawi. We assembled 94% of the 400 Mb genome, anchored 80% of the assembly to the 10 A. evenia chromosomes and predicted 32,667 protein-coding genes, providing a platform for comparative genomics and analysis of the nitrogen-fixing symbiosis in legumes.
A. evenia genome v1.0
| Analysis name |
Whole genome Assembly and Annotation of Aeschynomene evenia (GenoToul) |
| Date performed |
2018-03-03 |
| Consortium |
The Aeschynomene evenia reference genome sequence was completed and analyzed thanks to collaborative work between IRD (UMR LSTM, Montpellier, France), the GenoToul BioInformatics Facility (Toulouse, France) and CIRAD (UMR AGAP, Montpellier, France). |
| Family |
Fabaceae |
| Assembly |
This version (v1.0) of the assembly is 376 Mb (out of 400 Mb) spread over 1,848 scaffolds. 32,667 protein-coding genes were predicted, each with at least one primary transcript. Non-coding RNAs, repetitive elements and sequence variations were also analyzed. |
| Materials & Methods |
A total of 390 PacBio scaffolds (representing 302 Mb) was anchored based on GBS data and synteny analysis with Arachis spp. These scaffolds were joined to generate 10 pseudomolecules that were named according to the linkage group nomenclature. Each scaffold join was denoted with 100 N base pairs. 1,446 scaffolds (totalizing 73.8 Mb) remain unmapped in the current genome release and were grouped arbitrarily into a pseudomolecule named "Unknown”, each scaffold being joined by 100 Ns. |
| Reference |
Genetics of nodulation in Aeschynomene evenia uncovers mechanisms of the nitrogen-fixing rhizobium symbiosis
Johan Quilbé, Léo Lamy, Laurent Brottier, Philippe Leleux, Joël Fardoux, Ronan Rivallan, Thomas Benichou, Rémi Guyonnet, Manuel Becana, Irene Villar, Olivier Garsmeur, Bárbara Hufnagel, Amandine Delteil, Djamel Gully, Clémence Chaintreuil, Marjorie Pervent, Fabienne Cartieaux, Mickaël Bourge, Nicolas Valentin, Guillaume Martin, Loïc Fontaine, Gaëtan Droc, Alexis Dereeper, Andrew Farmer, Cyril Libourel, Nico Nouwen, Frédéric Gressent, Pierre Mournet, Angélique D’Hont, Eric Giraud, Christophe Klopp and Jean-François Arrighi.
Nature Communications. 2021 Feb 5;12(1):829. doi: 10.1038/s41467-021-21094-7.
|
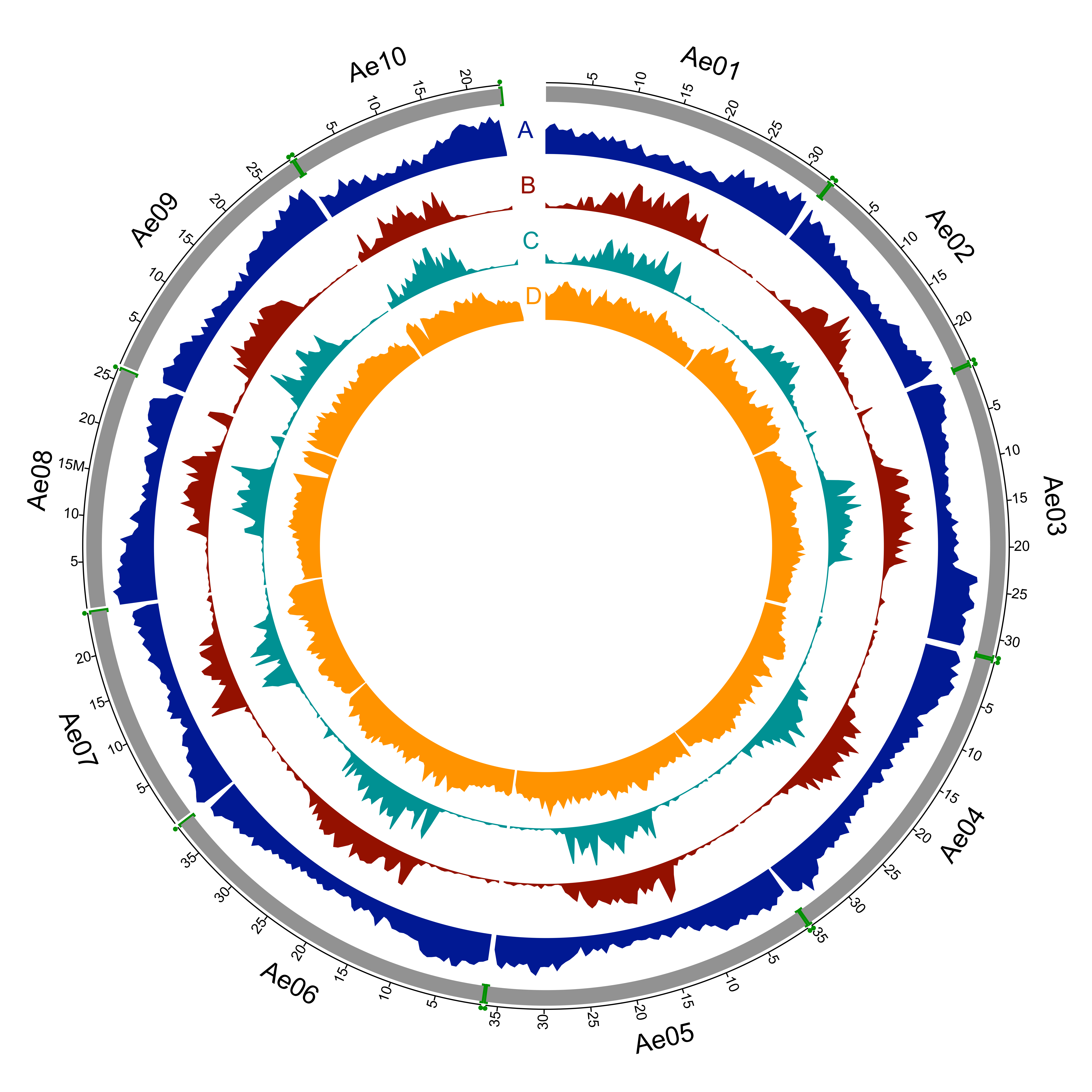
Gene Predictions
The following data types are currently present for this organism
| Feature type |
Count |
| Chromosome |
10 |
| Genes |
32,667 |
| Average gene length |
3,196 bp |
| Average CDS length |
1,221 bp |
| Average exons per gene |
5.04 |
|








